Research
Our research explores the interface between supramolecular chemistry, biology, catalysis and material science by studying dynamic combinatorial libraries: molecular networks involving dynamically exchanging chemical species. We study complex mixtures of interacting and interconverting molecules with particular focus on the new properties that can emerge from molecules acting collectively. We work in the emerging field of "Systems chemistry". Our work is relevant to disparate fields ranging from the origins of life (self-replicating systems, dynamic kinetic stability) to materials chemistry (self-synthesising fibres,gels and surface functionalized nanoparticles). Several projects are currently underway.
Systems chemistry
Traditionally chemists have been trained to work with molecules in isolation, i.e., pure compounds. Yet biological systems function by virtue of an overwhelming complexity of interlinked molecular processes. Using modern analytical techniques we have started the study of complex mixtures of synthetic molecules that can interconvert as well as interact noncovalently. Such mixtures constitute networks that can transmit molecular information. New properties can be expected to emerge from molecules acting in concert that are relevant to understanding how Nature evolved its complex molecular networks and, ultimately, to the origins of life. Furthermore, once we have learned how to design and control complex molecular systems, we should be able to design new functions complementary to those encountered in Nature, and eventually we should be able to synthesize life de-novo.
Self-replication: Towards de-novo life
In 2010, we reported in Science the spontaneous emergence of two different self-replicating peptide-derived macrocycles from a complex equilibrium mixture. Both replicators compete for an identical feedstock. Competition is influenced by the mode of agitation: shaking favours one replicator while stirring leads to dominance of the other one. The process of self-replication is driven by self-organisation of the replicators into nanofibres large enough to be observed by electron microscopy.
New building blocks functionalised with new peptides chains and other recognition motifs are now being synthesized in order to investigate the self-replicating behaviour of the system and therefore get a better understanding of the interactions within the system, which appears to be driven by an autocatalytic self-recognition process. We have also started to explore systems containing several building blocks and have recently observed exciting phenomena, including adaptation to changes in the environment and behaviour that resembles speciation as it occurs in biology.
Self-Synthesizing Materials
Notwithstanding the development of many synthetic replicating systems, studies of the primitive evolution has been hampered by the inherently limited variability of the replicators. Supramolecular systems such as this one, being able to perform templated synthesis of themselves without any need of enzymatic machinery, enable us to study origins of life pathways.
The self-templated DCLs described above also constitute a promising method for the development of new self-assembling materials. Self-assembly provides the driving force to pull the system away from equilibrium, in favor of the very molecules that self-assemble, so that these materials can in effect be deemed self-synthesizing.
As the fibres described above form through a nucleation-growth mechanism, it is possible to control fibre lenght and achieve length distributions with uniquely narrow polydispersities. These results are among the first examples of living supramolecular polymerization and allow for the first time, access to supramolecular block-co-polymers.
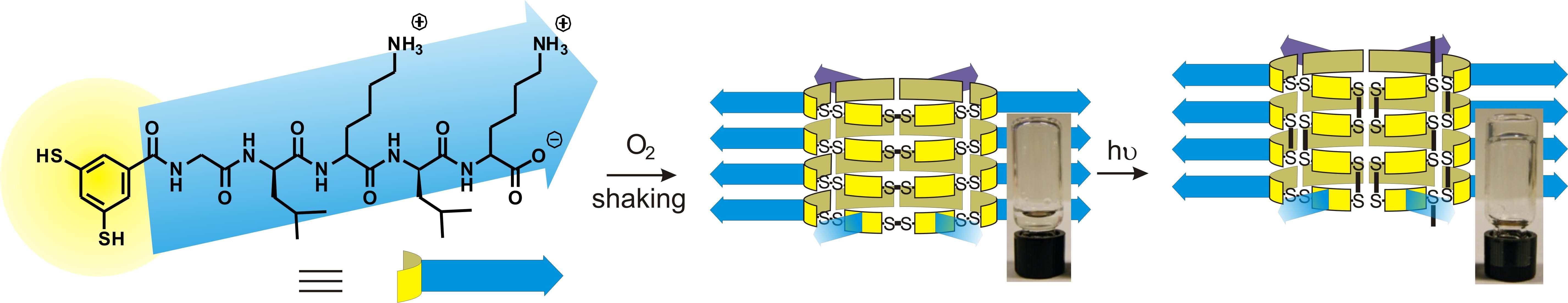
We found that the self-assembled fibers produced by dynamic combinatorial chemistry can be further stabilized by rearranging the dynamic covalent disulfide bonds that were underlying the dynamic combinatorial process. We showed how photo-initiated disulfide exchange converts fibrous stacks of macrocycles into polymeric products, enhancing the stability of the fibers and causing gelation of the aqueous solution.2
Receptors for biologically relevant targets from dynamic combinatorial libraries
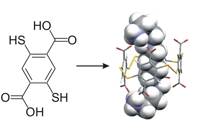
We have developed reversible disulfide chemistry to produce dynamic combinatorial libraries of macrocyclic receptors arising from a mixture of many possible subunits. Adding a guest to a dynamic library results in a shift of the equilibrium in favour of the best host, which can then be identified and isolated. Using this approach we have developed receptors for a wide range of guests. For example, we discovered a receptor with nanomolar affinity for spermine (H2N-(CH2)3-NH-(CH2)4-NH-(CH2)3-NH2) which can indirecty control the conformation of DNA. The building block from which the receptor is made and the structure of the host-spermine complex are shown below.
The concept of dynamic combinatorial molecular recognition has also been applied in the field of anion receptors. Nanomolar affinity and impressive selectivity result from a delicate balance between rigidity and flexibility that is achieved only thanks to the diversity that a dynamic combinatorial library can produce, out of which the best receptor can emerge, which structure would otherwise be difficult to predict.
Molecular recognition at biomembrane interfaces
While cell membranes play many vital roles in life, the important interactions occurring on/in cell membranes are still poorly understood. One of the main problems is that the insights into host-guest recognition in bulk solution often translates poorly to the lipid bilayer interface of the membrane.
Our approach to the problem is to develop Dynamic Combinatorial Chemistry (DCC) as a new tool to investigate molecular recognition at the bilayer interface, a phenomenon that is hard to study with currently existing methodologies. The use of DCC at the lipid bilayer interface is unprecedented and it will build the first bridge between the thus far separate areas of dynamic combinatorial chemistry and membrane chemistry and biophysics. Since the microenvironment of the lipid bilayer differs greatly from the bulk solution, it is likely that host-guest interactions will be very different in the two environments. This should lead to new insights into the effect of the special microenvironment of the lipid bilayer interface on molecular recognition.
Such insight is not only of fundamental importance to biology, but also essential for further development of medicinal applications of compounds that act at sites in the neighbourhood of biological membranes.
We intend to use these systems as models to study important biological processes like membrane transport, cell-cell recognition and ultimately processes like cell fusion and receptor-mediated endocytosis.
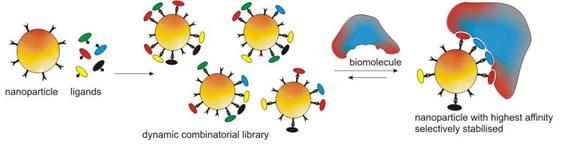
Dynamic Combinatorial Nanoparticles for Biomacromolecule Recognition
Combining the merits of dynamic combinatorial chemistry (DCC) as a tool for molecular recognition and nanoparticle systems renders a new approach to biomacromolecule detection by offering a platform with extended recognition surface and multivalent recognition. We aim to achieve highly specific surface functionalisation of hybrid nanoparticles through DCC. Nanoparticles decorated with functional groups that can participate in controllable reversible covalent bond formation have been prepared. Ligands as potential recognition units for biomacomolecules are then immobilized on the surface of nanoparticles through DCC. The recognition process is realized by exposing the equilibrium mixtures to the targets, which should lead to a shift in the product distribution towards the nanoparticles that have the highest affinity for the target molecules. We recently obtained the first very promising results using DNA as a template.
Towards Feedback Control over Catalysis in Dynamic Molecular Networks
This research area combines dynamic combinatorial chemistry in water with catalysis. The first step is to produce catalysts out of hydrazone or disulfide dynamic combinatorial libraries. In order for that to happen, the substrate should initially interact with the library so that a low-energy transition-state complex can be formed. If the product of the catalysis also interacts with the library members, feedback loops may emerge in the catalytic system. Our aim is to study these feedback loops and how they affect the rate of catalysis. We recently reported a dynamic molecular network in which the introduction of a substrate induced the transient formation of a catalyst that converts this substrate. After the substrate has been consumed the catalyst dissappears by re-equilibrating into other library members.
Selected publications:
- Z. Rodriguez-Docampo, E. Eugenieva-Ilieva, C. Reyheller, A. M. Belenguer, S. Kubik, S. Otto, P. Herdewijn
Dynamic combinatorial development of a neutral synthetic receptor that binds sulfate with nanomolar affinity in aqueous solution.
Chem. Comm. 2011, 47, 9798-9800. doi: 10.1039/C1CC13451E. - J. Li, J. M. A. Carnall, M. C. A. Stuart, S. Otto
Hydrogel formation upon photoinduced covalent capture of macrocycle stacks from dynamic combinatorial libraries.
Angew. Chem., Int. Ed. 2011, 50:36, 8384–8386. doi: 10.1002/anie.201103297. - F. M. Mansfeld, H. Y. Au-Yeung, J. K. M. Sanders, S. Otto
Dynamic combinatorial chemistry at the phospholipid bilayer interface.
J. Syst. Chem. 2010, 1:12. doi: 10.1186/1759-2208-1-12. - J. M. A. Carnall, C. A. Waudby, A. M. Belenguer, M. C. A. Stuart, J. J.-P. Peyralans, S. Otto
Mechanosensitive self-replication driven by self-organization.
Science 2010, 327, 1502-1506. doi: 10.1126/science.1182767. - R. F. Ludlow, S. Otto
Systems chemistry.
Chem. Soc. Rev. 2008, 37, 101-108. doi: 10.1039/B611921M. - P. T. Corbett, J. K. M. Sanders, S. Otto
Systems chemistry: pattern formation in random dynamic combinatorial libraries.
Angew. Chem., Int. Ed. 2007, 46, 8858-8861. doi: 10.1002/ange.200702460. - L. Vial, R. F. Ludlow, J. Leclaire, S. Otto
Controlling the biological effects of spermine using a synthetic receptor.
J. Am. Chem. Soc. 2006, 128, 10253-10257. doi: 10.1021/ja062536b. - P. T. Corbett, J. Leclaire, L. Vial, K. R. West, J.-L. Wietor, J. K. M. Sanders, S. Otto
Dynamic combinatorial chemistry.
Chem. Rev. 2006, 106, 3652-3711. doi: 10.1021/cr020452p. - B. Brisig, J. K. M. Sanders, S. Otto
Selection and amplification of a catalyst from a dynamic combinatorial library.
Angew. Chem. Int. Ed. 2003, 42, 1270-1273. doi: 10.1002/anie.200390326. - S. Otto, R. L. E. Furlan, J. K. M. Sanders
Selection and amplification of hosts from dynamic combinatorial libraries of macrocyclic disulfides.
Science 2002, 297, 590-593. doi: 10.1126/science.1072361.